Genomics
GEBV-assisted breeding to accelerate breeding of Chile pepper with improved disease resistance, pungency, quality, and adaptation. This theme focuses on utilizing genomic estimated breeding values (GEBVs) derived from genomic prediction models as selection criteria in advancing lines and selecting parents for the Chile pepper breeding program at New Mexico State University. GEBV-based breeding for more pungent (hot), disease resistant, high-quality, and more adapted Chile pepper varieties will be implemented. The quantitative nature of these traits will make genomic prediction strategies relevant in the breeding program. For example, the measure of heat level in Chile pepper, called Scoville Heat Units (SHU), could be predicted in lines that are yet to be evaluated in experimental trials using genomic selection models. Various ridge regression and Bayesian models and schemes will be explored and implemented, and prediction accuracy achieved from these selection models will be evaluated. Furthermore, the effects of different parameters such as training population size, composition, relatedness between training and validation populations, number of markers, and genetic architecture on prediction ability will be examined. In the context of disease resistance in Chile pepper, another research avenue that could be further explored would be the use of Function Associated Specific Trait (FAST) SNP markers (Fu et al. 2017) from RNA-sequencing, which has not been conducted for genomic prediction of resistance to Phytophthora capcisi (Barchenger et al. 2018). Gains from selection achieved from this approach will be compared to genetic gains from traditional phenotyping and other selection strategies.
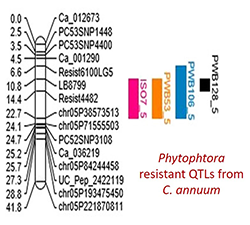

Dissecting the genetic basis of important yield, quality, and disease resistance traits in Chile pepper through association mapping, QTL validation, and candidate gene analyses. GWAS will be implemented to identify genomic regions controlling important fruit quality, yield, and disease resistance traits of Chile pepper varieties grown in New Mexico. The implementation of molecular marker-assisted breeding for disease resistance has been successful primarily due to the presence of relatively few QTLs with major effects controlling these traits (Hospital 2009). In New Mexico, one of the major diseases in Chile pepper is root rot (Chile Wilt) and pod rot caused by the soilborne fungus Phytophthora capcisi (Goldberg 1995). A major focus of Theme 2 will be the identification of SNP markers associated with these diseases to facilitate the development of resistant lines through marker-assisted selection strategies. Recently, genomic regions associated with root rot have been identified in pepper chromosomes P5, P7, and P11 using genotyping-by-sequencing (GBS)-based QTL mapping and GWAS approaches. Three major effect QTLs in P5 were observed to be related to broad-spectrum resistance to P. capcisi (Siddique et al. 2019). In another study, a single region on chromosome P5 was also found to be related to disease resistance in four Phytophthora isolates in bell pepper (Chunthawodtiporn et al. 2019; see figure on the left). Molecular markers in the form of competitive allele-specific (KASP®) assays from these previously identified QTLs, particularly for the major effect loci in chromosome P5, could be developed to validate their effects on existing Chile pepper germplasm at New Mexico State University. Once validated, these KASP assays could be used routinely in the breeding program to accelerate the selection of genotypes with improved disease resistance to soilborne diseases. The functions of genes underlying these traits will be further identified through candidate gene analyses to gain a broader and deeper understanding of the genetic architecture of these diseases.